Single-cell Amplified Genomes
Background
90 % of the ocean’s biomass comprise of marine microorganisms where the SAR11 bacteria is the most ubiquitous, comprising of 30 % of all surface water cells and one of the most widespread organisms on Earth. Its ability to harvest labile dissolved organic matter and produce volatile organic compounds plays an important role in global biogeochemistry. Its small genome size and complexity provides insight into the microbiological evolution and function of streamlined cells. However, complete SAR11 whole genomes sequences are relatively rare due to their difficulty in culturing, leading to gaps in knowledge of SAR11 diversity. Here we show the advantage of single-cell amplified genomes in genomic analysis of the SAR11 clade. 451 SAR11 bacteria were isolated and sequenced using SAG technology. We found evidence suggesting the presence of two new phylogenetic clades of SAR11, expanding the numbers of niche habitats SAR11 inhabits. Additionally, genomic analysis confirms the existence of multiple hypervariable regions containing niche-defining genes. Analysis of these HVRs revealed genes related to nutrient uptake and phage defense, consistent with previous studies advocating for a King of the Mountain ecological strategy. Lastly, identification of viral signatures within SAR11 SAGs lead to confirmation of existing viral clades within SAR11. Our results demonstrate the value of SAGs to allow for clade wide studies of the SAR11 pangenome. We anticipate the usage of SAGs to become a powerful alternative for the study of difficult to isolate and culture organisms.
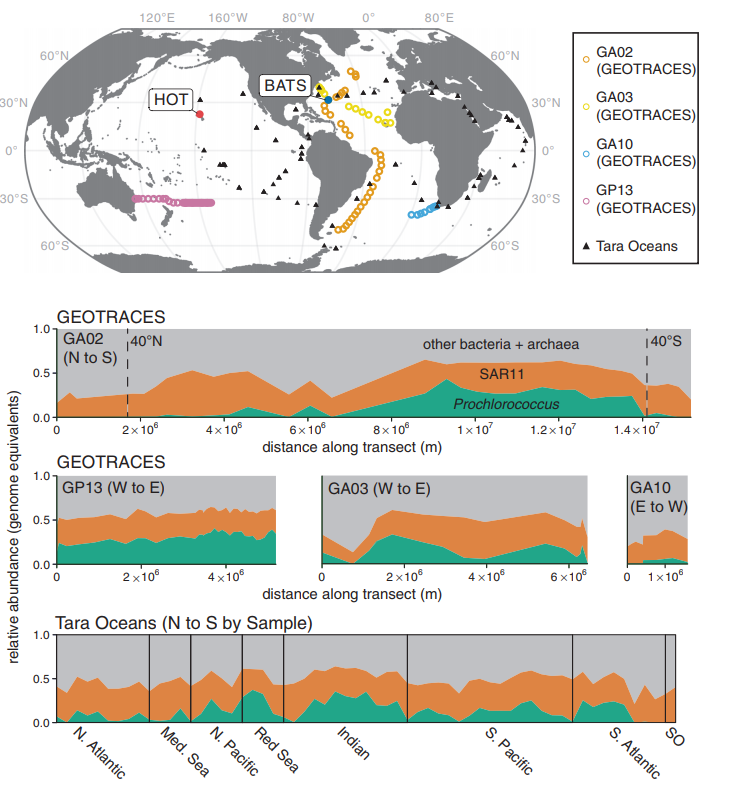 Becker et al. 2019 Fig 1. Co-culture and biogeography of Prochlorococcus and SAR11
Becker et al. 2019 Fig 1. Co-culture and biogeography of Prochlorococcus and SAR11
Single-cell Amplified Genomes
Single-cell Amplified Genomes (SAGs) are genomes isolated from single cell sorting techniques and are whole genome amplified and sequenced. Organisms usually have only 1 or a few copies of their genomic DNA, which is not enough to sequence. Amplification of this genomic DNA allows for pure samples of an organism to be sequenced without the need for culturing. This is particularly useful for organisms that are difficult to grow or are slow growing. 451 SAR11 SAGs are sequenced within this study. SAR11 is an aquatic bacteria and the most abundant microbe in the world’s oceans. Culturing this slow growing organism is difficult and time consuming. SAG technology provides an alternative to traditional culting or metagenomic studies of these ubiquitous organisms.
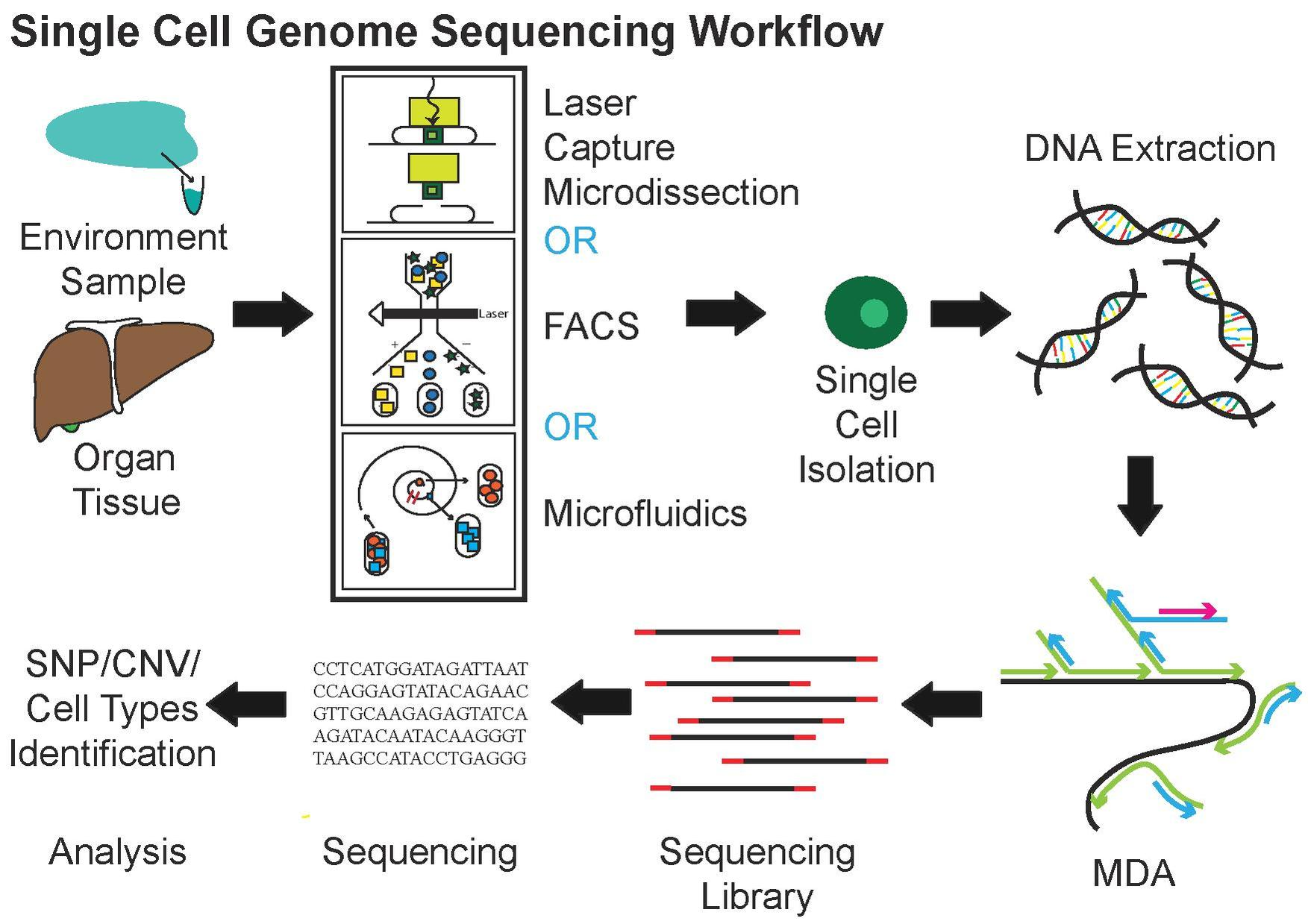 Wikipedia Single Cell Sequencing
Wikipedia Single Cell Sequencing
451 SAR11 SAGs
SAGs were assembled and phylogentics determined to allow for catagorisation of SAGs into existing SAR11 phylogentic clades. Viral signatures within SAGs were extracted and analysed and hypervariable regions determined based on metagenomic data.
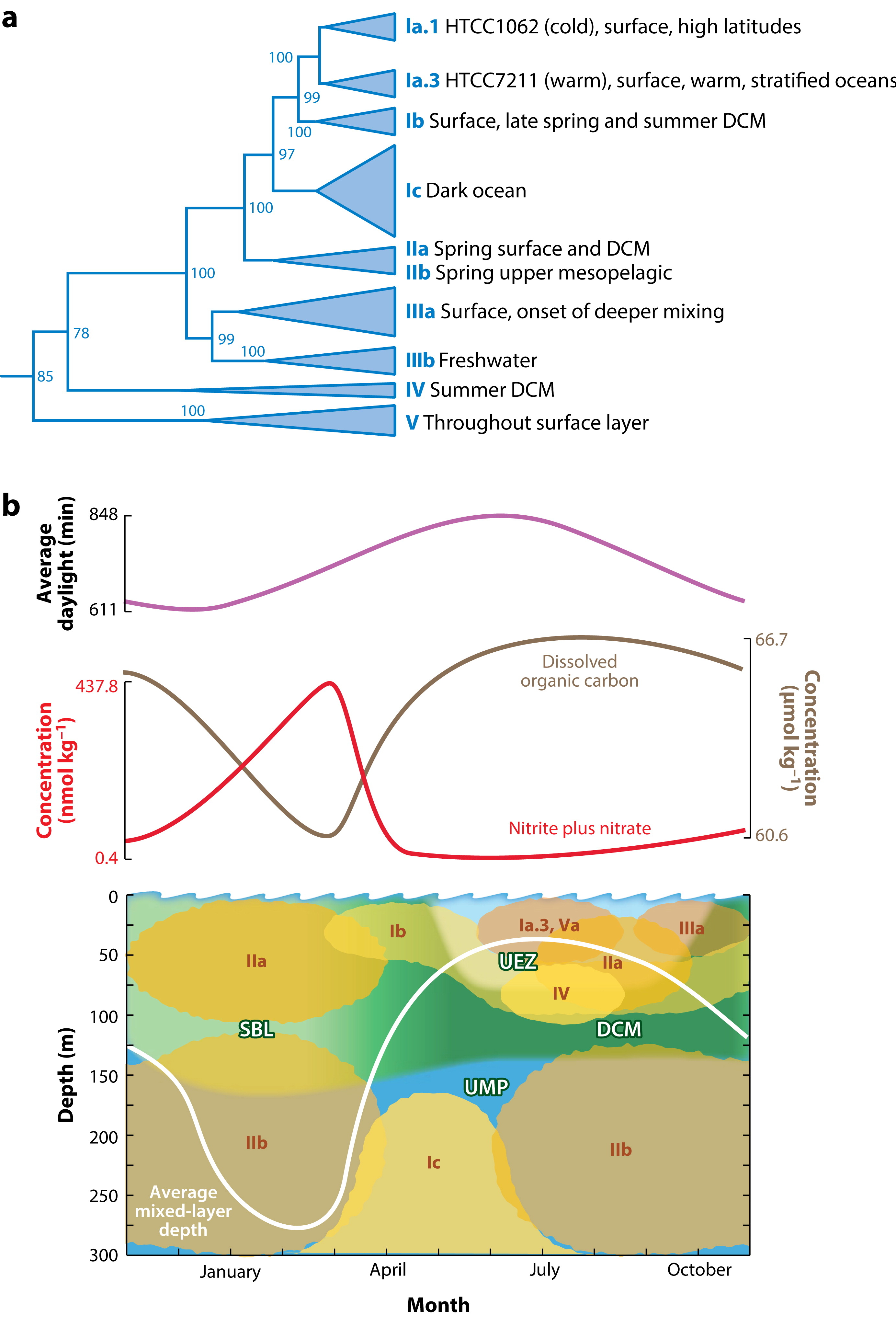 Giovannoni 2017 (a) Phylogenetic tree of SAR11 diversity, showing the major ecotypes. (b) Spatiotemporal distribution of the SAR11 ecotypes at the BATS site.
Giovannoni 2017 (a) Phylogenetic tree of SAR11 diversity, showing the major ecotypes. (b) Spatiotemporal distribution of the SAR11 ecotypes at the BATS site.
Phylogenetic Tree
Generally, phylogenetics refers to how related a species is to another based on some kind of simularity, like their genetic content. Similarly grouped species sit within the same “branch” of a phylogenetic tree, with more distantly related species placed further away. To establish relatedness, conserved genes of all species like 16/18S rRNA genes are used as points of comparison. This is because all cells have a form of 16/18S rRNA gene within their genome. This allows for comparisons of similarity between such individuals.
Within this study, phylogenetics is used to first describe the relationship of each SAG in comparison to each other. This would allow for categorisation of each SAG into the established 5 main clades of SAR11. Average Amino acid Identity AAI was used to confirm splits within the branch structure of phylogenetic trees. Metagenomic data of differing spatial and temporal locations within the ocean were then used to deduce if splits within phylogentics trees corrispond with any ecological niches.
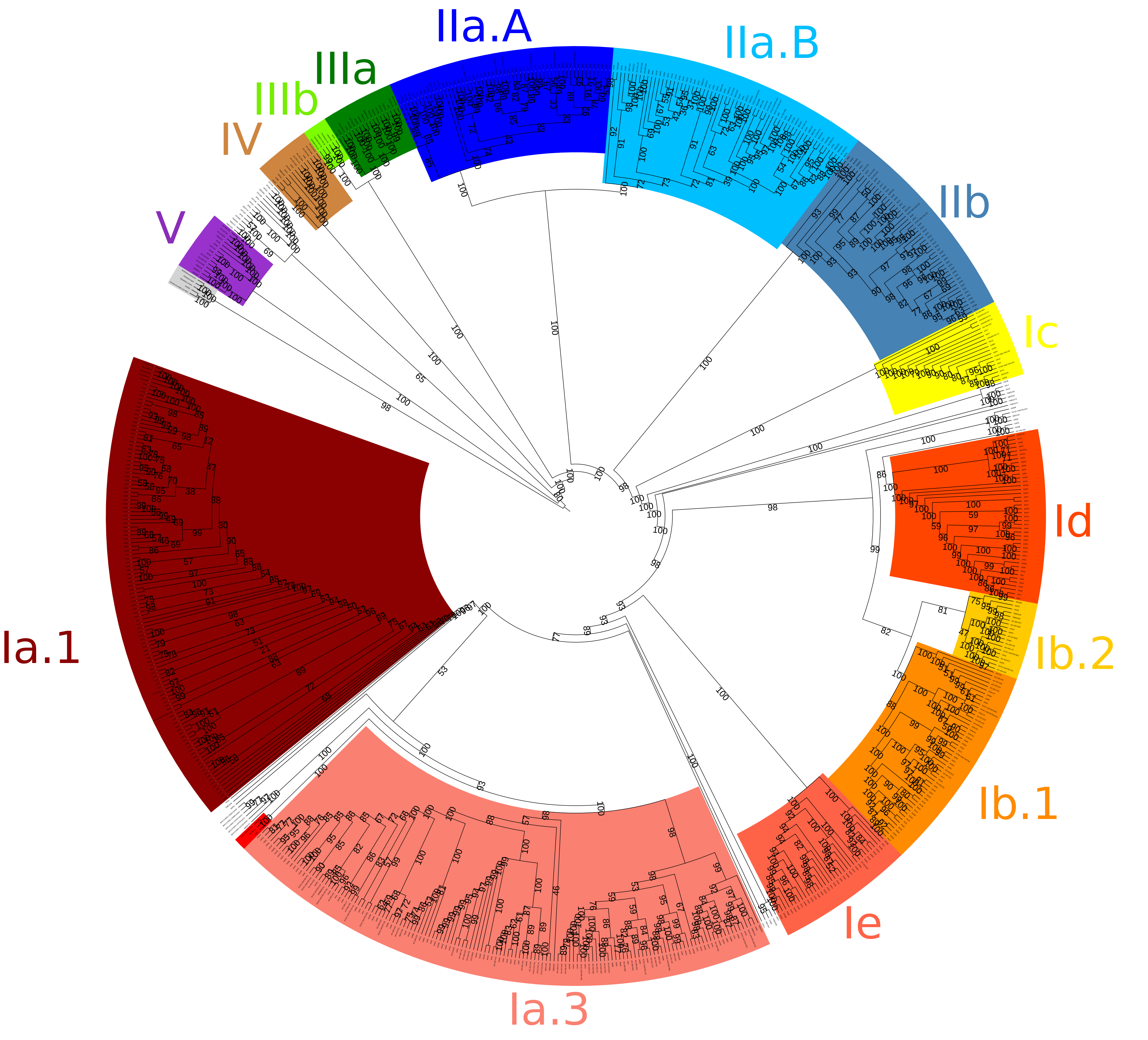 Phylogenetic Tree of SAR11 SAGs and reference genomes divided by clades
Phylogenetic Tree of SAR11 SAGs and reference genomes divided by clades
Viral Signatures
It has long been established that viruses have a large impact on ecosystems. They are key factors in regulating bacterial and eukaryotic microorganism populations. They can infect both multicellular and single cell organisms often with detrimental effects to the host. Within single-cell organisms, they are responsible for the turnover of over 20 % of marine bacteria biomass daily and contribute enormously to the differentiation of the number of genes an organism may have through integration of temperate phages. Any study looking at single cell populations must consider the population of viruses within the same ecosystem.
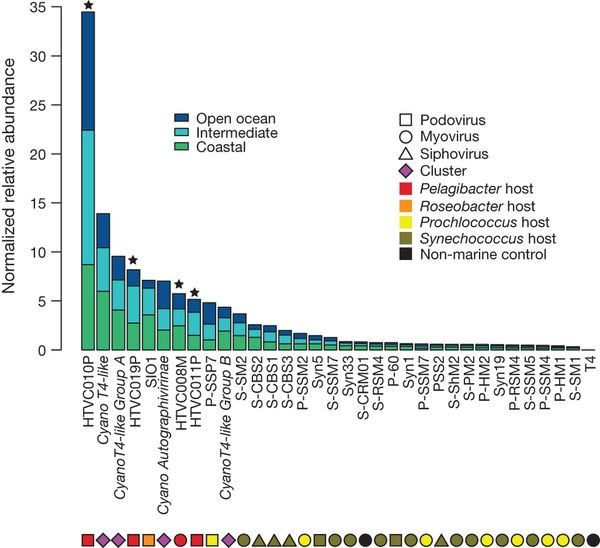 Zhao et al 2013 Figure 3. Abundance of pelagiphage relative to representative cyanophages and roseophage SIO1
Zhao et al 2013 Figure 3. Abundance of pelagiphage relative to representative cyanophages and roseophage SIO1
In 2013 it was discovered that there were abundant SAR11 viruses in the ocean contrary to established ecological theories related to kill the winner hypothesis. As a result, SAR11 SAGs were analysed computationally for viral signatures. 21 viral signitures of over 10 kbps were extracted and analysed. Results indicate a close relationship with existing SAR11 phages based on shared gene content as well as phages related to other marine viruses but not SAR11 related.
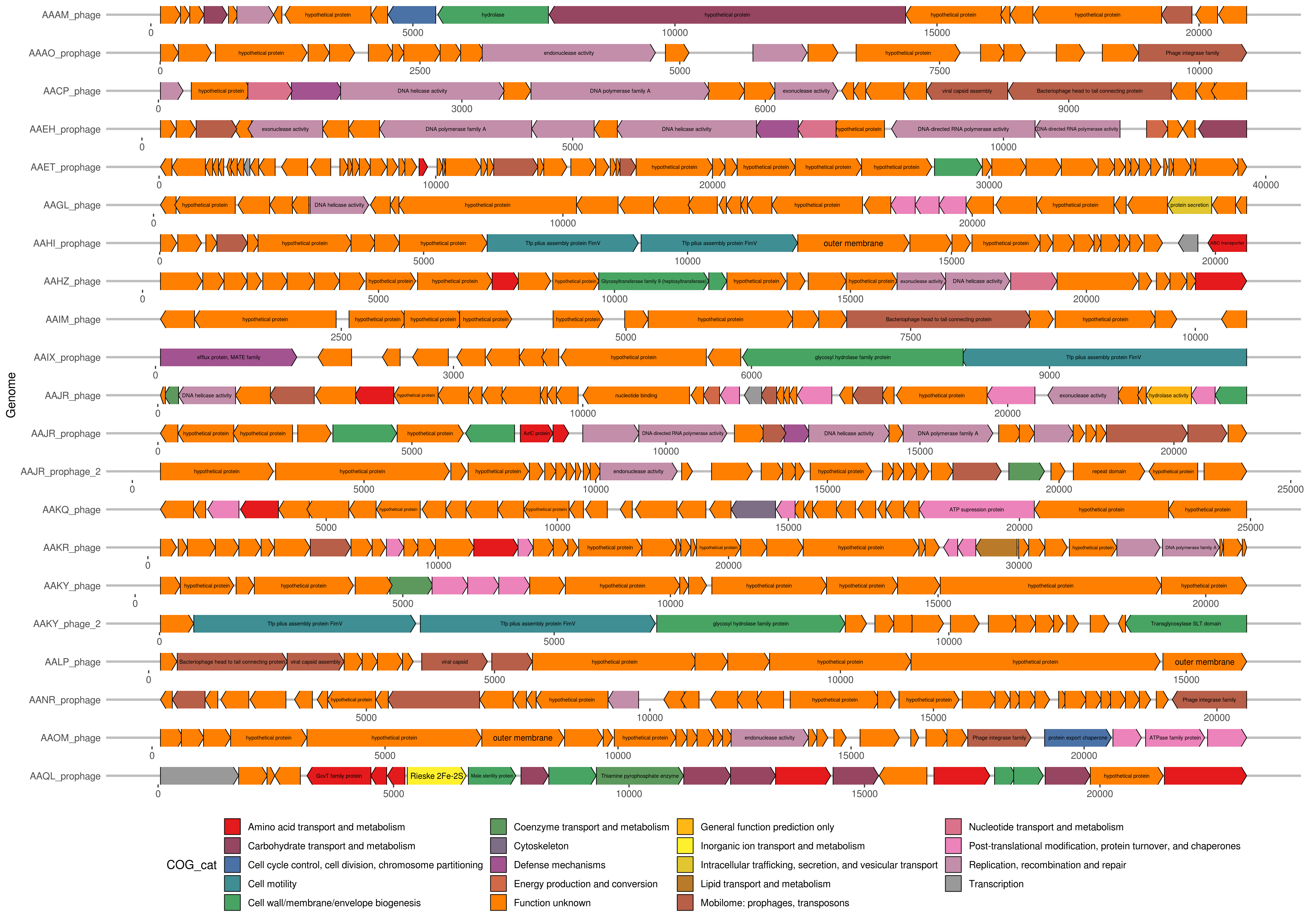 21 SAR11 SAG viral gene annotations with colours representing COG functional groups
21 SAR11 SAG viral gene annotations with colours representing COG functional groups
Hypervariable Regions
Hypervariable Regions (HVRs) are genomic regions undergoing higher evolutionary rates in comparison to other areas of its genome. This allows for genes located within these regions to undergo higher mutation rates resulting in new and novel gene formations or slight variations in existing genes that conferr additional fitness.
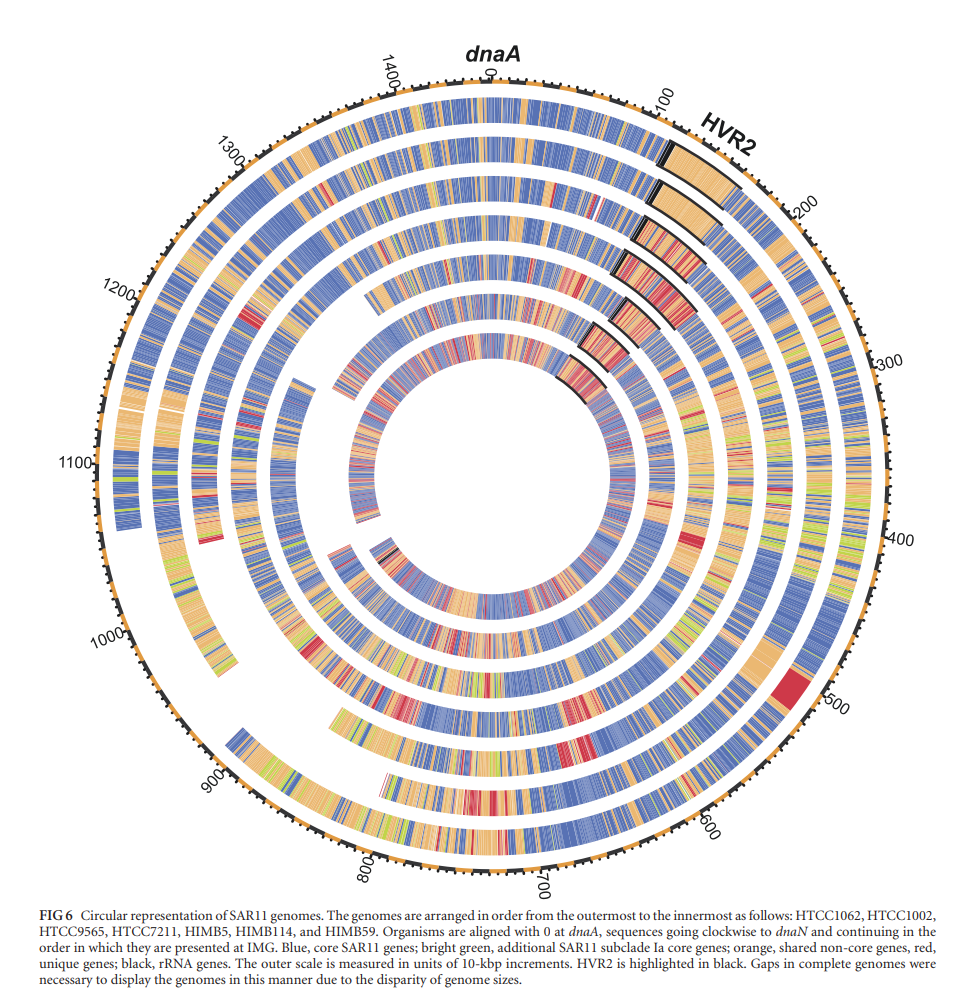 Grote et al 2012 Fig 6. Streamlining and Core Genome Conservation among Highly Divergent Members of the SAR11 Clade
Grote et al 2012 Fig 6. Streamlining and Core Genome Conservation among Highly Divergent Members of the SAR11 Clade
SAR11 has been hypotheised to contain HVRs conferring fitness in nutrient uptake and phage defense. With a highly streamline genome, rates of homolgus recombination occur at one of the highest known rates. This would allow for regions conferring fitness in genes related to phage survial and nutrient uptake to be passed quickly between individuals and evolved in parallel. This would support the current King of the Mountain ecological strategy proposed to explain how the SAR11 clade thrives with high abundance of phages and in areas of low nutrients. Pangenomic analysis of all SAR11 HVRs indicate an gene enrichment of genes related to cell wall/membrane function.
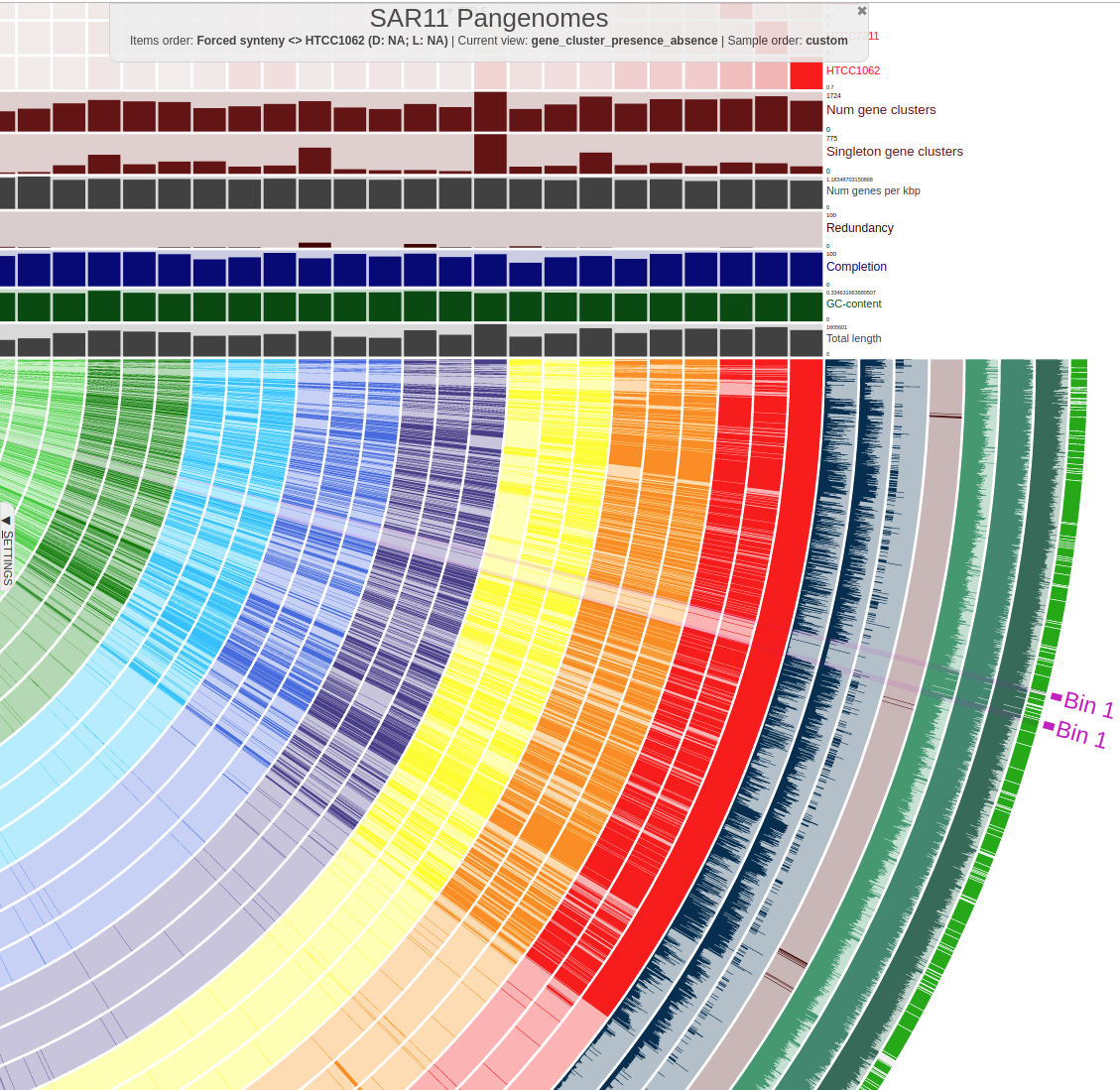 SAR11 HVR flanking genes conserved throughout all SAR11 clades visualised by anvi’o
SAR11 HVR flanking genes conserved throughout all SAR11 clades visualised by anvi’o
