Marine Metagenomics
Intro
With the world’s oceans covering roughly 70 % of the earth surface; its impact on the global carbon cycle, climate change, and acid-base biochemistry remains crucial to our understand of the natural world. Considering 90 % of the ocean’s biomass comprise of marine microorganisms; their impact on global systems remain largely understudied. Marine microorganisms are critical in the energy cycle and are the foundation for marine life. However, studying these microorganisms remains challenging with a small fraction being culturable for in situ experimentation. Alternative study methods include obtaining genomes via metagenomics studies and Single-cell Amplified Genomes. Both of these methods have advantages and drawbacks with marine metagenomes being highly complex to analyse and SAGs suffering from both low coverage and bias. This study aims to explore both approaches and provide improvements to aid analysis.
Metagenomics
Metagenomics is the study of any genetic content recovered from an environmetal sample, usually used to describe population dynamics, gene content and to recover genomes from organisms that are challenging to culture. Historically, organisms were isolated in the laboratory, cultured and DNA was harvested for sequencing. With metagenomics, samples are collected and sequencing without isolation of any specific organism. This allows for entire populations to be captured within a sample. Metagenomes can also be assembled to produce multiple whole genomes of organisms using specalised computer algorithims. However, increasing numbers of similarly related organisms within a sample pose higher conplexity and challenges to obtain whole genomes sequences.
Visualisation
During metagenomic assembly, we attempt to reconstruct whole genomes from short reads. However, these are often fragmented and steps are required to try and group together pieces of genomes that are thought to belong to the same organism. This can be done by visualising the nucleotide composition of the sequences and identifying those that are similar. Here contigs - sequences of genetic code that share similar nucleotide composition profiles are plotted on a scatter plot via a t-SNE technique for visualisation. Colours are overlayed over contigs based on their phylogentic content.
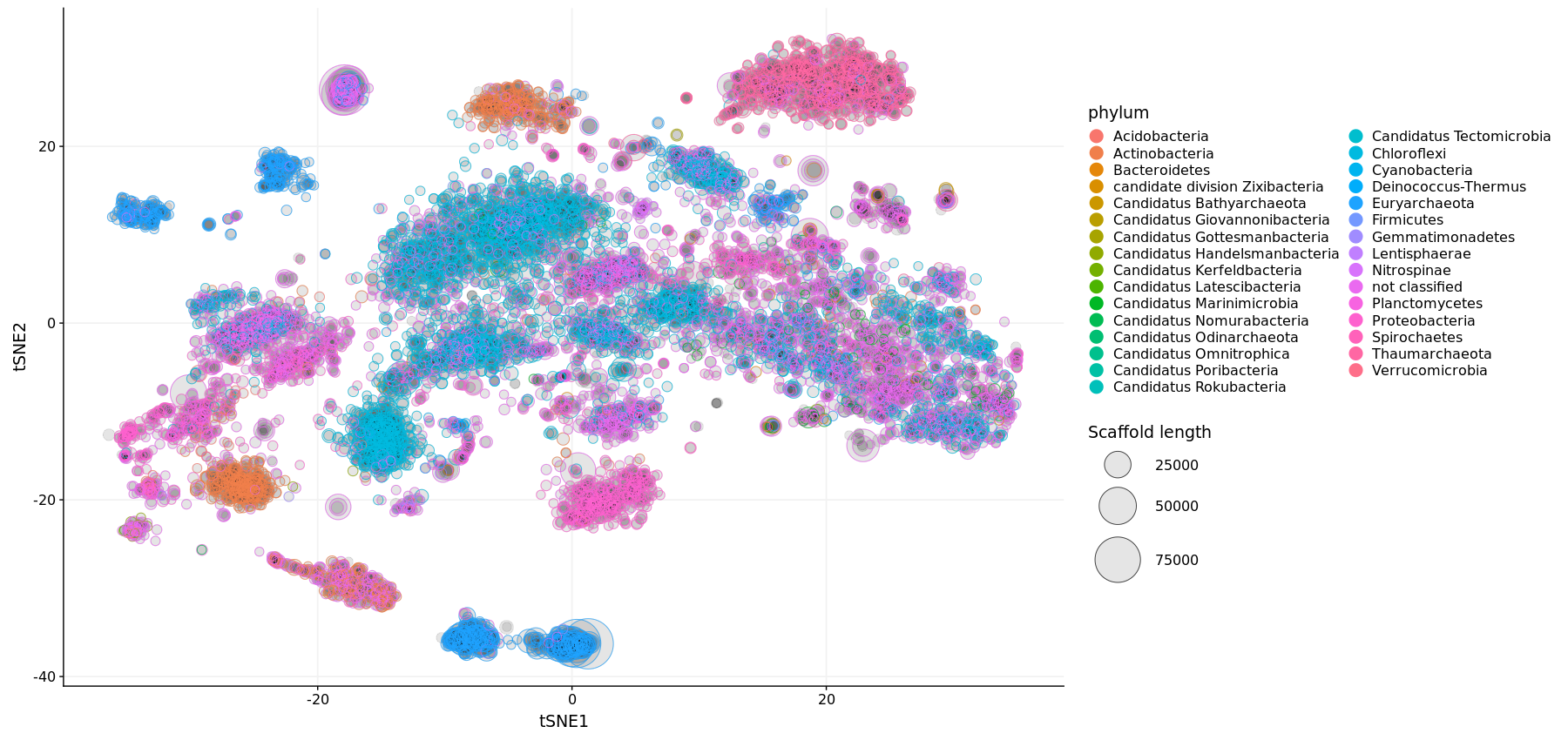
Evalution of t-SNE plots on recovering marine metagenomic samples reveal limited sucess. SAR11 genomes were increasing prone to fragmentation wereas other none marine organisms clustered based on k-mer counting.
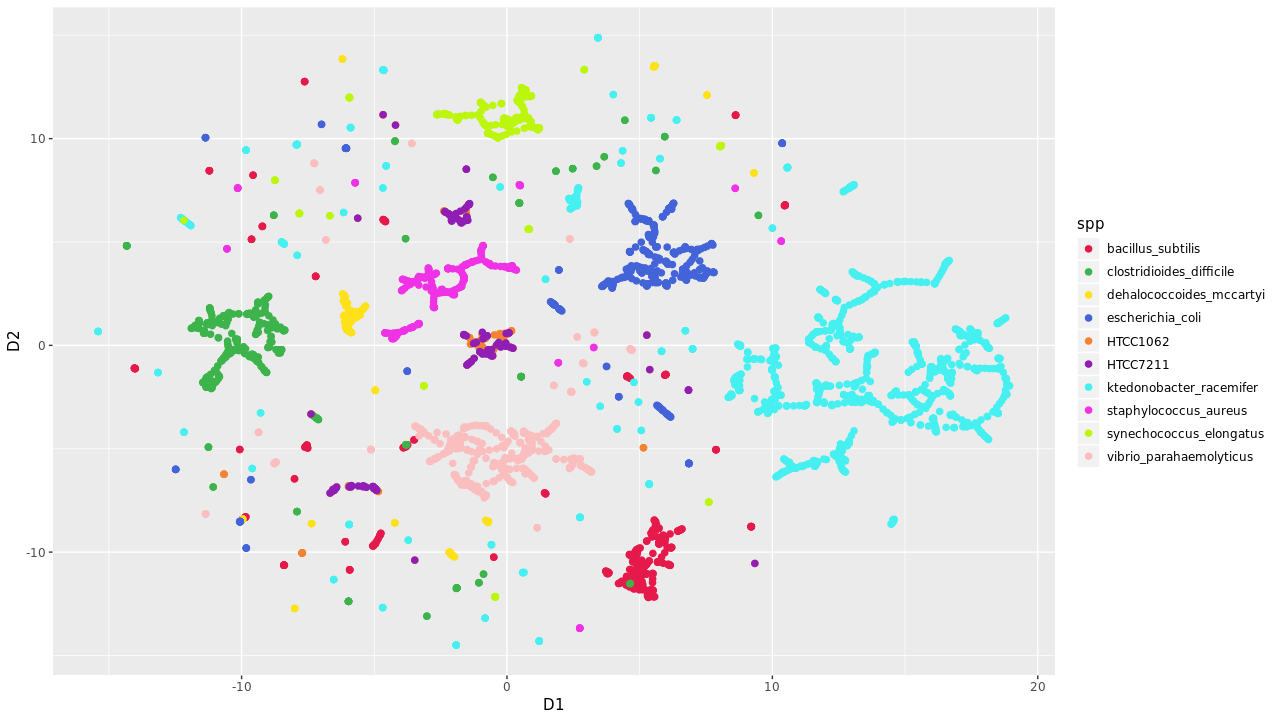
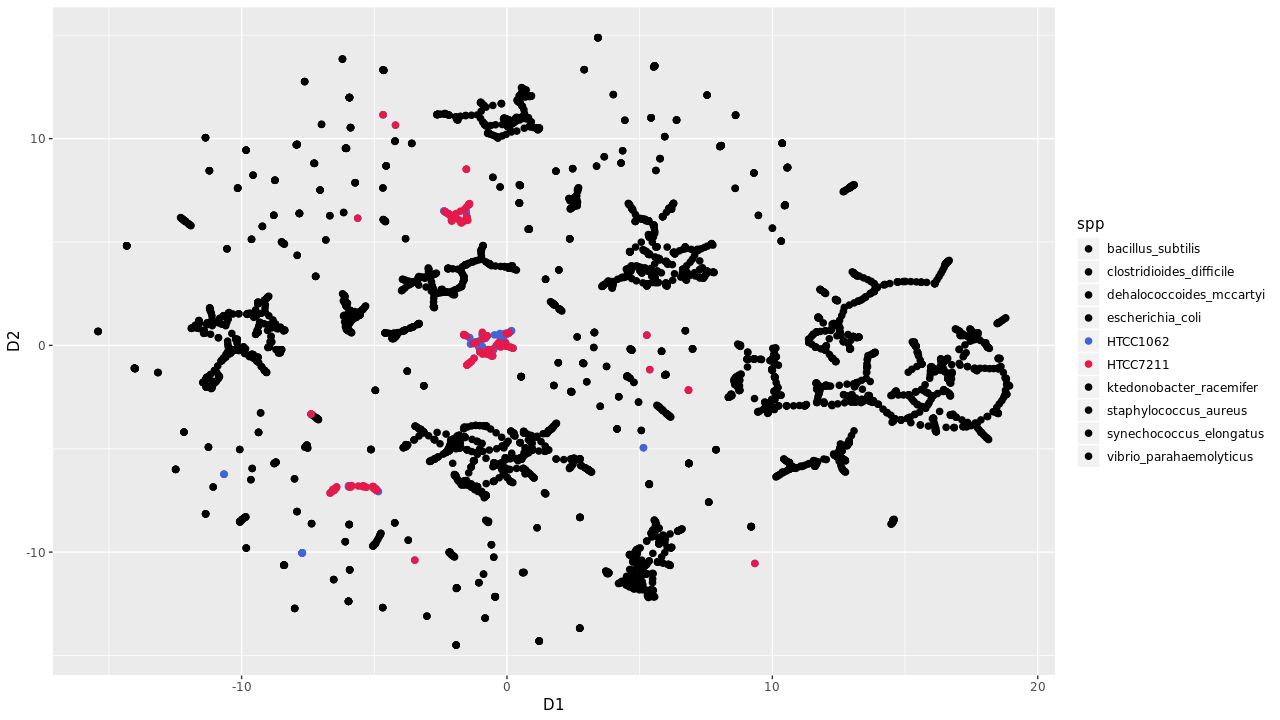
Global Abundance
With metagenomes, all genetic components within a sample are sequenced, allowing for a snapshot of a microbial population with a certain space and time. Mapping or comparing an exisiting isolate to a metagenome can reveal the abundance of that isolate within that community. This provides a mechanism to predict sample presence absence or relative abundance of taxa within a large timeframe and spatial zone, without the need to take new genetic samples. Below is an example of SAR11 Single-cell Amplifed Genome abundance, a dominant marine microorganism at differing times and depths at the Bermuda Atlantic Time Series, and its isolated corresponding bacteriophage.
